The model of the syser was proposed independently by V.A. Ratner
and V.V. Shamin, D.H. White, and R. Feistel [1-3]. .
A syser includes a polynucleotide matrix I,
a replication enzyme E1, a translation
enzyme E2 , and optional proteins E3
, E4, ..., En
(Fig. 1).
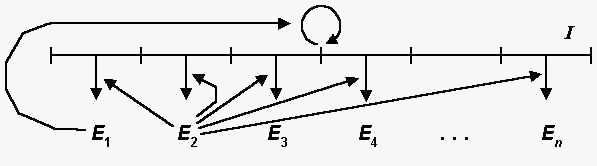
Fig 1. The general scheme of a
syser. I is the polynucleotide matrix, E1
and E2 are the replication and the
translation enzymes, respectively, E3 ,
E4, ..., En
are optional proteins.
The polynucleotide matrix I codes
proteins, the replication enzyme E1 provides
the matrix replication process, the translation enzyme E2
provides the protein synthesis according to an information,
stored in the matrix I . We can imply that there is the
translation enzyme system (consisting of several enzymes) rather
than the single translation enzyme - such a substitution does not
change the mathematical description of sysers. The same is valid
for the replication enzyme.
Whereas hypercycles
[4] can be treated as a plausible model of the origin of translation mechanisms, sysers are more similar to real biological organisms
than hypercycles. Nevertheless, possible scenarios of an
origin of simple sysers from small molecules were also discussed
[1,2]. Contrary to the hypercycle, the syser has a universal RNA
replication enzyme. The mathematical analysis of sysers [3,5,6]
is similar to that of hypercycles.
Analogously to hypercycles, different sysers
should be placed into different compartments for effective
competition. For example, we can model the sysers' competition,
using the following assumptions [7]: 1) the different sysers are
placed into separate coacervates [8], 2) any coacervate
volume grows proportionally to the total macromolecules synthesis
rate, 3) any coacervate splits into two parts when its volume
exceeds a certain critical value. During a competition, a syser,
having a maximal total macromolecules synthesis rate, is selected
[3,5].
The model of sysers provides the ability to
analyze evolutionary stages from a mini-syser, which contains
only matrix I and replication E1 and
translation E2 enzymes, to protocell, having
rather real biological features. Some features can be modeled by
assigning certain functions to optional proteins (Fig.1). For
example, "Adaptive syser"
[6] includes a simple molecular control system, which "turns
on" and "turns off" synthesis of some enzyme in
response to the external medium change; the scheme of this
molecular regulation is similar to the classical model by F. Jacob
and J. Monod [9]. The mathematical models of the adaptive syser as
well as that of the mini-syser are described in the child node Adaptive syser: the model of hypothetical
prebiological control system .
The scheme of sysers is similar to that of the
self-reproducing automata by J. von Neumann [10]. The
self-reproducing automata components and their syser's
counterparts can be represented as follows:
Self-reproducing automata by J. von Neumann
|
Sysers
|
Linear storing chain L
|
Polynucleotide matrix I
|
Constructing automaton A for
manufacturing an arbitrary automaton according to
description, stored in the chain L
|
Translation enzyme E2
|
Automaton B for copying of the
chain L
|
Replication enzyme E1
|
Automaton C, needed to control
the whole reproduction and to separate of the
produced "child" system from the
"parent" one
|
Splitting of a coacervate during a syser growth
|
Conclusion. Sysers is a rather
universal model of self-reproducing system. It provides the
ability to analyze evolutionary stages from a very simple
macromolecular systems to protocells, having real biological
features.
References:
1. V.A. Ratner and V.V. Shamin. In:
Mathematical models of evolutionary genetics. Novosibirsk: ICG,
1980. P.66. V.A. Ratner and V.V. Shamin. Zhurnal Obshchei
Biologii. 1983. Vol.44. N.1. PP. 51. (In Russian).
2. D.H. White. J. Mol. Evol. 1980.
Vol.16. N.2. P.121.
3. R. Feistel. Studia biophysica.1983.
Vol.93. N.2. P.113.
4. Eigen M. and P. Schuster. The Hypercycle: A principle of natural self-organization, Springer, Berlin, 1979
5. V.G. Red'ko. Biofizika. 1986. Vol. 31.
N.4. P. 701 (In Russian).
6. V.G. Red'ko. Biofizika. 1990. Vol. 35.
N.6. P. 1007 (In Russian).
7. R. Feistel, Yu. M. Romanovskii, and
V.A.Vasil'ev. Biofizika. 1980. Vol. 25. N.5. P. 882. (In
Russian).
8. A.I. Oparin. "The origin of
life". New York, 1938. A.I.Oparin. "Genesis
and evolutionary development of life". New York,
1968.
9. F. Jacob and J. Monod. J. Mol. Biol.
1961. Vol. 3. P. 318.
10. von Neumann J. Theory of Self-Reproducing Automata. (Ed. by A. W. Burks), Univ. of Illinois Press, Champaign, 1966.

